Data in MLatom’s Python API
Published Time: 2024-08-14 16:06:28
Data is cornerstone of MLatom as it is a data-driven package. To truly unlock the potential of MLatom, you need to master handling of data with its Python API.
In our online tutorial, we give a primer into MLatom’s data and you will learn, for example, how to view normal mode vibrations or MD in Jupyter with a single line.
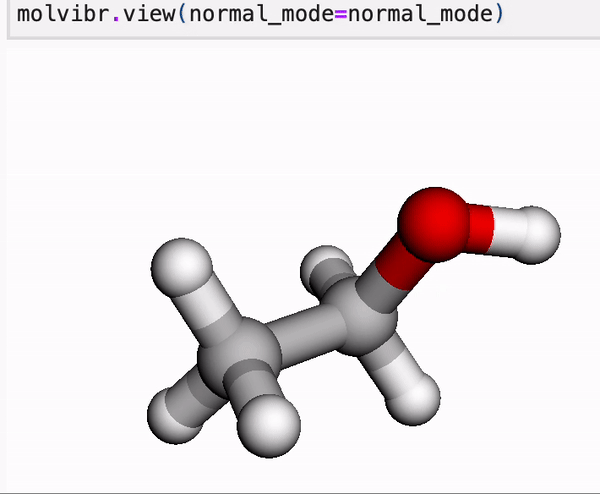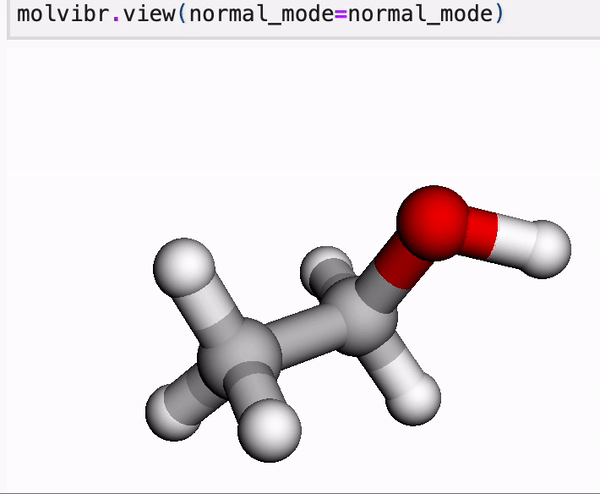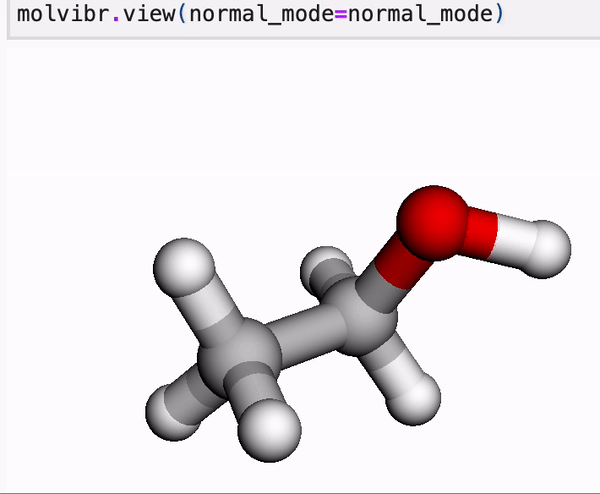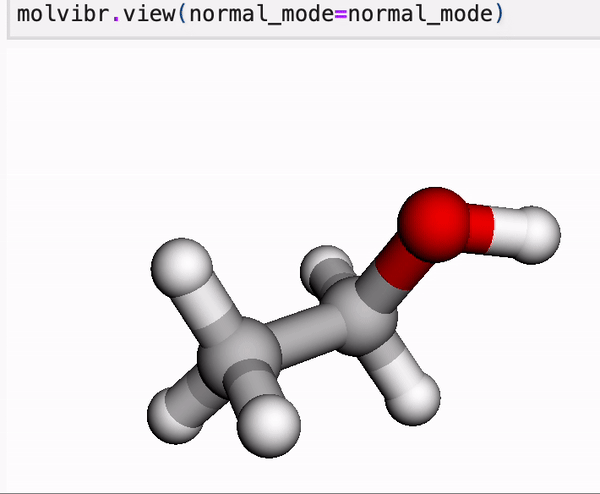
Molecule is a central concept in MLatom as most of operations are usually done on molecule class objects. These objects store information about the constituent atom objects, their coordinates, and any property we want to learn or calculate. Molecules can be loaded and dumped in various formats.
In machine learning, we need data with many molecules. MLatom for this uses the molecular_database class. The databases can be loaded and dumped in different formats. The useful feature of molecular databases is that they can be manipulated as lists/numpy arrays or split into several other databases. This is useful, for example, when we need to split the data into training and test sets.
Molecular dynamics trajectories are handled with a dedicated molecular_trajectory class. The trajectory step contains the usual information about the molecule, step number, time, energies, and so on.
Just to mention that if you are dealing with the excited states and surface hopping trajectories, MLatom has an intuitive access to complex properties through molecule.electronic_states.
__________________________________________________________________________________________________________________________________
If you want to learn more about AI-enhanced computational chemistry, you can check out our new hands-on online mini-course, perfectly suitable for both beginners and experts wanting to upgrade their computational tools.