Introduciton to XEDA in XACS
Published Time: 2022-06-29 09:49:09
We recently released the Xiamen Atomistic Computing Suite (XACS), which consists of three core software packages:
(1) XMVB – A Valence Bond Theory Based Quantum Chemistry Program
(2) XEDA – A General and Multipurpose Energy Decomposition Analysis Program
(3) MLatom – A Package for Atomistic Simulations with Machine Learning
In our previous post, we introduced XMVB software and will now introduce the second package in the suite – XEDA.
Molecular interactions play a very important role in molecular structures and properties. As one of the main approaches to quantitatively study molecular interactions, energy decomposition analysis methods can be widely used in many fields, such as drug design, molecular self-assembly, chemical reaction mechanisms, acidity/alkalinity, aromaticity, and molecular force fields.
Xiamen energy decomposition analysis program, XEDA, has been developed by Prof. Peifeng Su and his students for more than 10 years. XEDA is the only EDA program, which can analyze various intermolecular interactions, ranging from weak interactions (van der Waals, hydrogen bonds) to strong covalent bonds (triple bonds), from gas phase to condensed phase, from closed-shell to open-shell molecules, from ground to excited states.
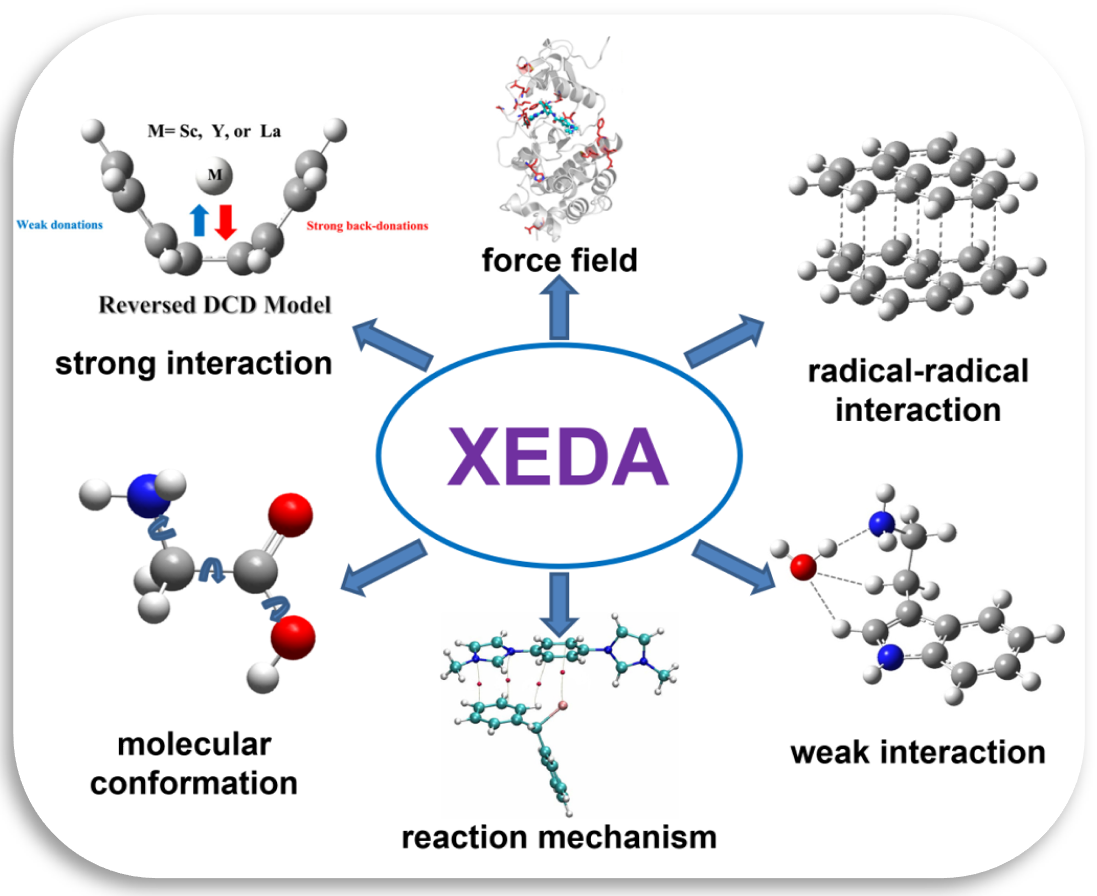
XEDA employs LMO-EDA, GKS-EDA and their extensions to explore the physical origin of intermolecular interactions. The memory and time consumptions are the same level of single point energy calculations. Because of its accuracy, efficiency and capability for complex systems, GKS-EDA method has been widely used. Please see the review (WIREs Comput. Mol. Sci. 2020; e1460) for the details.
There are two ways to access XEDA package and use it for calculations: most conveniently online using our cloud computing platform and also locally after downloading and installing the package. This post shows how to perform XEDA calculation on our cloud computing platform.
1. Starting off
With XACS cloud computing, users do not need to download or install software packages. You can simply visit the XACS official website (http://xacs.xmu.edu.cn), register for free or login to your existing account, submit calculations, track the progress and retrieve the calculation results.
2. Example
Take the GKS-EDA calculation of adenine and thymine base pair as an example, which is available on the XACS website (http://xacs.xmu.edu.cn/records?example_id=108&editable=0&attribution_program=XEDA). Here is the input file.

In this input file, the charge and multiplicity of each monomer, along with the type of EDA method, are assigned in $LMOEDA. Especially, edatyp=gks shows the calculation is performed with GKS-EDA.
Visit the XEDA cloud computing page and copy the above input file into the text box (shown above). Click Submit to start the XEDA calculation. During the calculation, click “my computing” at the bottom of the page to track the progress of the job in time.
Once the calculation is complete, you can find a .png file, which is the visualized EDA results as below.

This figure shows the EDA results for the interaction between adenine and thymine base pairs. Negative values represent attraction while positive values show the repulsive contribution to the total interaction. Among these attraction terms, electrostatic term is the largest and polarization term takes the second. It is noted that the contribution of correlation and dispersion (corr/disp) cannot be neglected.
Welcome to visit the XACS website for more help. If you have any questions, please contact us via xmvb@xmu.edu.cn.
XACS homepage:http://xacs.xmu.edu.cn
XEDA manual:http://xacs.xmu.edu.cn/softwares/XEDA_manual.pdf
XEDA tutorial:http://xacs.xmu.edu.cn/softwares/XEDA_tutorial_2022.pdf
XEDA example:https://xacs.xmu.edu.cn/examples?attribution_program=XEDA